Sex is arguably a great invention.
It is, otherwise we could not acknowledge its existence across the tree of life: if sexual reproduction was not favoured by natural selection in at least some points in time and space, we would not observe it, this post would be about something that lacks a definition, and ultimately I would not even be alive. Instead, I will assume a strong consensus about sexual reproduction having an important role in evolution.
But why it is so?
I was prompted to think more deeply about this question when I started reading The selfish gene, the extremely popular book by Richard Dawkins. I remember some of my best professors from my Bachelor’s mentioning it, but I never even read the first chapters until recently (thanks to Yuyao for recommending it).
I know close to nothing about the many forms in which sexual reproduction occurs in the known species, but at the core of it it’s the generation of new genomes by the mixing of existing genomes. This is different from asexual reproduction, where new genomes are generated by directly duplicating existing ones.
There are convincing arguments supporting the idea that the most recent common ancestor of sexually reproducing species and asexually reproducing species was an asexually reproducing organism. Meiosis seems to be built on top of the molecular machinery that makes mitosis happen, but the additional complexity - involving the pairing of homologous chromosomes, the extensive recombination between non-sister chromatids, the suppression of sister chromatid separation during the first meiotic division, and the absence of chromosome replication during the second meiotic division - is staggering. Meiosis is indeed slower and more resource demanding than mitosis. Furthermore, the successful completion of meiosis is still not enough to make a new individual. The genome in a gamete has 0.5n ploidy and needs another gamete to generate a new individual. This merging step is yet another process, implying additional metabolic costs and chances that something could go wrong. In the meantime, the most joyful and carefree of asexual organisms might have gone through many cell cycles, successfully spreading its genes in the environment, each genome protected by an efficient, sex-free cell.
Of course, the fact that sexual reproduction exists despite being dramatically inefficient with respect to asexual reproduction has to be explained by some advantages inherent to the reshuffling of the genes in the gene pool of the parents to generate a new individual, as opposed to simple clonal expansion. This advantage has at least to match in some conditions the costs of a slower and more metabolically expensive cellular process.
Is this really true? What are the necessary conditions to observe the spreading of a novel “meiosis” trait (here used to indicate the ability to sexually reproduce) in a population composed of mainly mitotic organisms? How the conferred advantages look like? What are their properties? What is the contribution of crossing-over alone?
To try and answer these questions I adopted the ultimate tool for those who can not reach conclusions by theoretical reasoning… I ran a simulation.
Simulation
The environment in which the genomes evolve has the following parameters:
c: carrying capacity, i.e. the maximum number of genomes that can be concurrently sustained by the environmentn: number of genes in the evolving genomess: number of genomes that exist at the start of the simulationh_mean: mean number of genes between two crossing-over pointsh_sd: standard deviation of the number of genes between two crossing-over pointsx: starting fraction of genomes that can sexually reproduceg: number of generations the simulation will runm: mutation rate, i.e. the probability of an allele to mutate in the next generationq: the number of new individuals that a single mitotic organism will generate when reproducingu: the number of new individuals that a couple of meiotic organisms will generate when reproducing
The following assumptions hold:
- The genome is constituted by only one chromosome
- Genes are ordered, meaning that a distance between two genes A and B can be calculated in terms of how many genes separate A and B
- Each gene has two alleles. One allele is supposed to have no effect on the fitness, while the other can be beneficial or detrimental
- The genes’ effects on the fitness are independent and additive
- The effect of allele dosage is additive
- A genome is initialized randomly, each gene having a random dosage of the alleles
- The additional cost associated with sexual reproduction can be
simulated by tuning the parameters
qandu, affecting how many individuals are generated by an asexual or by a sexual reproduction event, respectively
Initialization
At the beginning of the simulation, s individuals are initialized randomly, each having a random genome conferring a random fitness. A (small) fraction x of these individuals has the meiosis trait.
Iterations
The simulation runs for g generations. In brief, during each of these iterations the following is applied:
- Compute the fitness of each individual in the current generation
- Rank the individuals from the highest to the lowest fitness, generating the propagation list
- Initialize an empty population for the new generation
- While the number of individuals in new generation is lower than the carrying capacity
c:- Remove the first individual parent A from the propagation list
- If parent A does not have the meiosis trait:
- Duplicate parent A
qtimes, generatingqidentical new individuals - Apply the mutation rate
mindependently to each of the new individuals - While the number of individuals in new generation is lower than the carrying capacity
c, add one by one the new individuals to new generation
- Duplicate parent A
- Else:
- Remove a second individual parent B from the propagation list that is the one with highest fitness among those with the meiosis trait
- If parent B exists, then
- Perform meiosis (perform crossing-over, generate gametes) in parent A and in parent B
- Randomly combine the gametes from parent A and parent B to create new individuals. These individuals have the meiosis trait
- While the number of individuals in new generation is lower than the carrying capacity
c, add one by one at mostuof these new individuals to new generation
Results
Below is displayed a run of the simulation. Results are very robust across many runs, but I still have to sistematically assess the simulation behavior across varying parameters, averaging on a large number of runs, and investigate the effect of individual mechanisms, e.g. what happens when we switch crossing-over off. Maybe, it will be the material for another post.
Black: meiosis trait
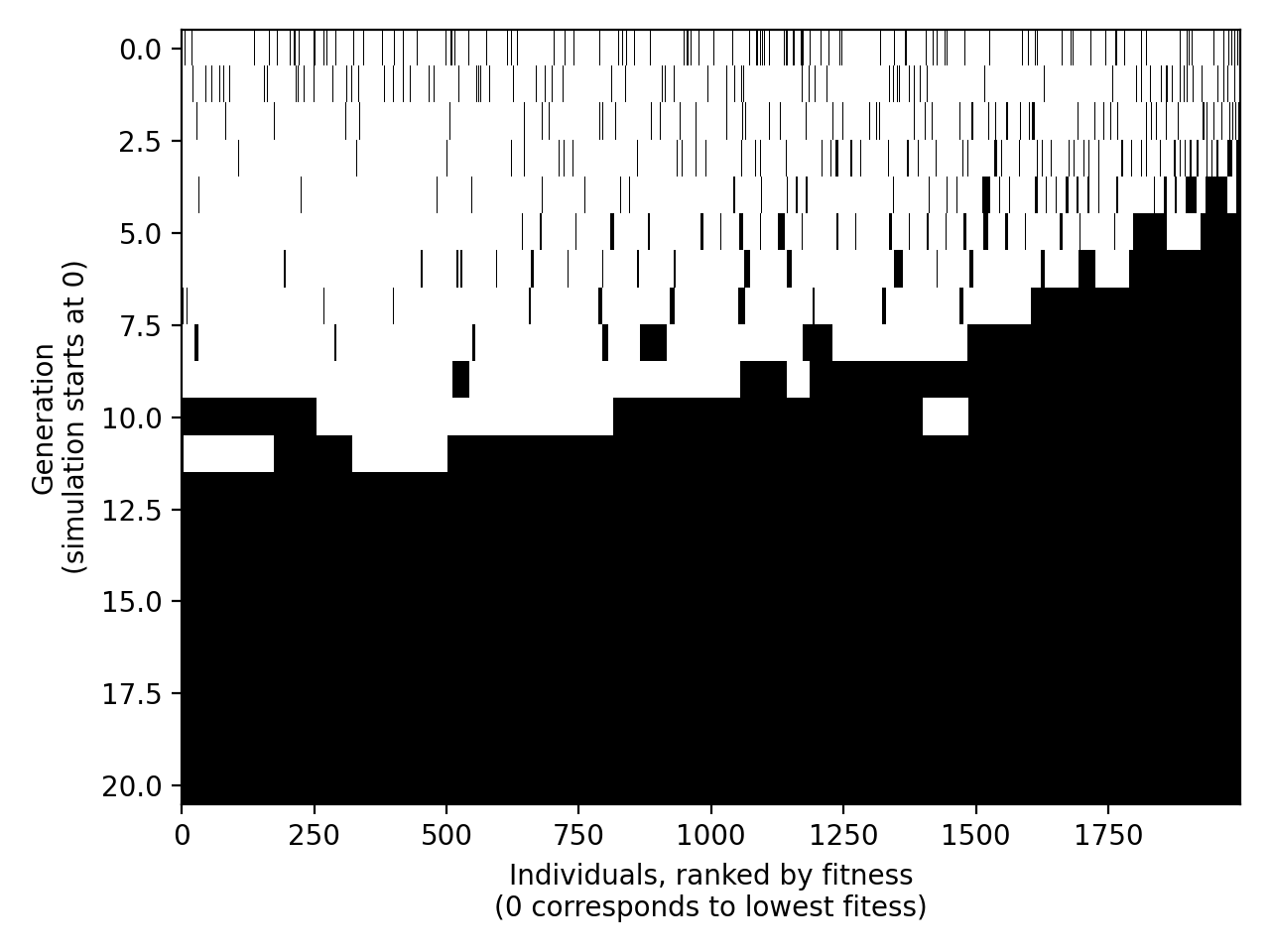
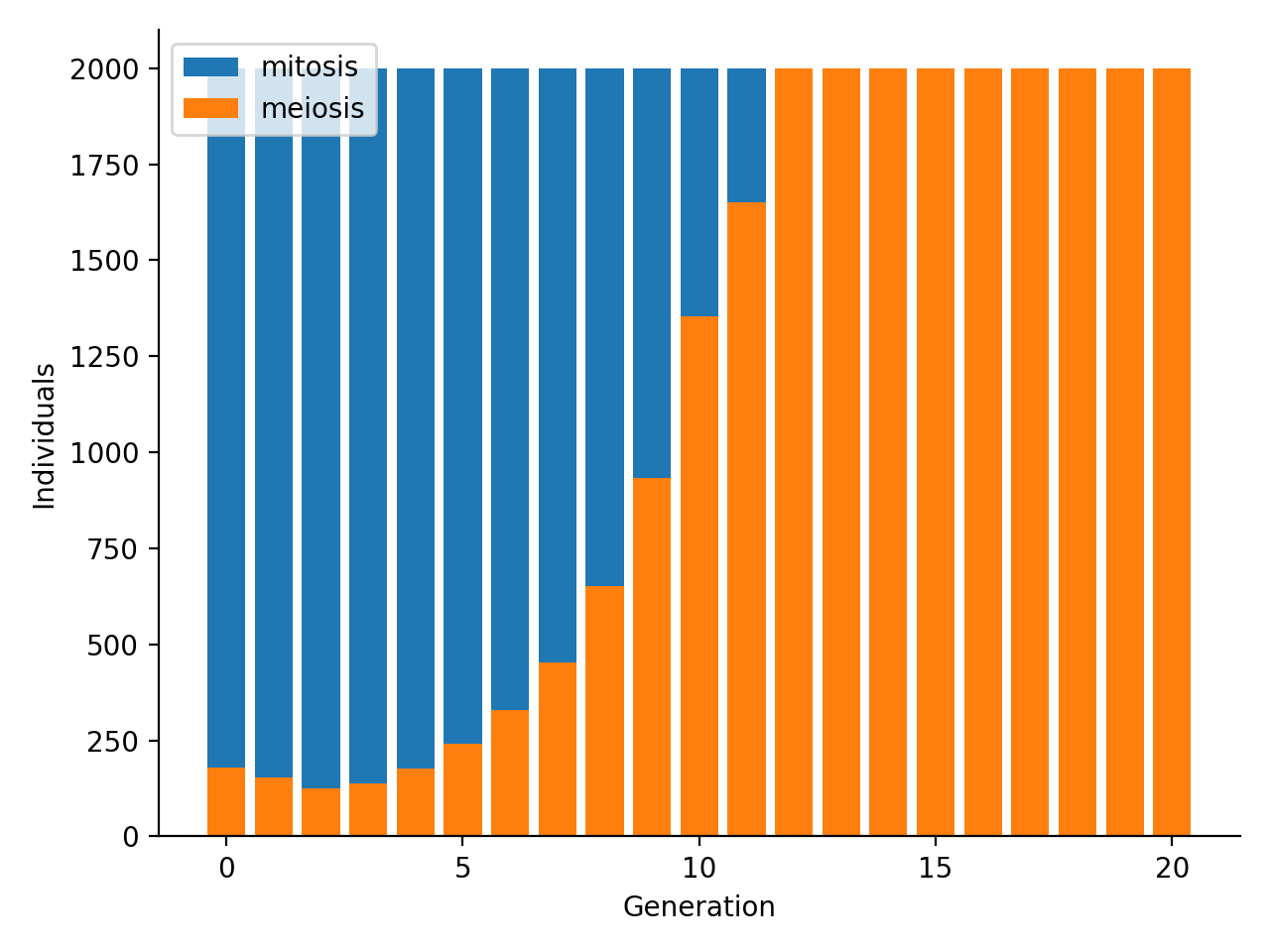
After briefly decreasing intially, the frequency of the meiosis trait explodes until reaching fixation in the population. If the individuals capable of sexual reproduction are not wiped out in the very first few generations because of the additional fitness cost linked to sexual reproduction (see parameters u vs q), their offspring will outcompete their non-sexually reproducing counterparts over the long run. It appears that genetic recombination greatly enhances the efficiency of exploration of the genotype/fitness space with respect to random mutations alone: at generation 0 of the simulation, sexually reproducing individuals are uniformly distributed on the fitness scale (upper figure), while in the following generations they 1) tend to form clusters towards the highest fitness ranks, and 2) their frequency in the population overall increases (lower figure).
Code
Source code and default parameters used here can be found at https://github.com/alussana/sex_emergence_simulation